Introduction
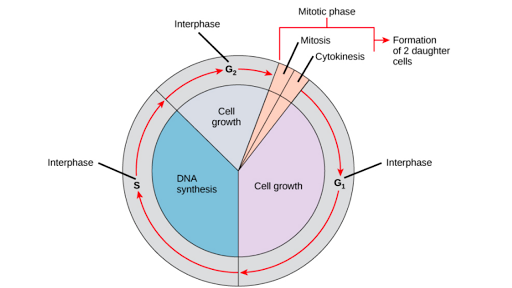
A cell cycle is a series of events that take place in a cell as it grows and divides. A cell spends most of its time in what is called interphase, and during this time it grows, replicates its chromosomes, and prepares for cell division. The cell then leaves interphase, undergoes mitosis, and completes its division. The resulting cells, known as daughter cells, each enter their own interphase and begin a new round of the cell cycle.
Gentaur Cell Cycle is the ordered sequence of events that occur in a cell in preparation for cell division. The cell cycle is a four-stage process in which the cell increases in size (gap 1, or G1, stage), copies its DNA (synthesis, or S, stage), prepares to divide (gap 2, or G2, stage), and divides (mitosis, or M stage). The G1, S, and G2 stages from interphase represent the time between cell divisions. Based on the stimulating and inhibitory messages a cell receives, it “decides” whether to enter the cell cycle and divide.
Proteins that play a role in stimulating cell division can be classified into four groups: growth factors, growth factor receptors, signal transducers, and nuclear regulatory proteins (transcription factors). For a stimulating signal to reach the nucleus and “activate” cell division, four main steps must occur. First, a growth factor must bind to its receptor on the cell membrane. Second, the receptor must be temporarily activated by this binding event. Third, this activation must stimulate the transmission or transduction of a signal from the receptor on the cell surface to the nucleus within the cell.
Finally, transcription factors within the nucleus must initiate the transcription of genes involved in cell proliferation. (Transcription is the process by which DNA is converted to RNA. Proteins are then made according to the RNA blueprint, and thus transcription is crucial as the initial step in protein production.) Cells use special proteins and checkpoint signalling systems to ensure that the cell cycle progresses correctly. Checkpoints at the end of G1 and the beginning of G2 are designed to assess DNA damage before and after the S phase. Likewise, a checkpoint during mitosis ensures that the cell’s spindle fibres are properly aligned in metaphase before the chromosomes separate in anaphase.

If DNA damage or spindle formation abnormalities are detected at these checkpoints, the cell is forced to undergo programmed cell death or apoptosis. However, the cell cycle and its checkpoint systems can be sabotaged by defective proteins or genes that cause malignant transformation of the cell, which can lead to cancer. For example, mutations in a protein called p53, which normally detects DNA abnormalities at the G1 checkpoint, may allow cancer-causing mutations to bypass this checkpoint and allow the cell to escape apoptosis.
Cell cycle stages
To divide, a cell must complete several important tasks: It must grow, copy its genetic material (DNA), and physically divide into two daughter cells. Cells perform these tasks in a series of organized and predictable steps that make up the cell cycle. The cell cycle is a cycle, rather than a linear pathway because, at the end of each round, the two daughter cells can start the exact same process from the beginning. In eukaryotic cells or cells with a nucleus, the stages of the cell cycle are divided into two main phases: interphase and the mitotic (M) phase.
- During interphase, the cell grows and makes a copy of its DNA.
- During the mitotic (M) phase, the cell separates its DNA into two sets and divides its cytoplasm, forming two new cells.
M phase
During the mitotic (M) phase, the cell splits its copied DNA and cytoplasm to form two new cells. The M phase involves two distinct processes related to division: mitosis and cytokinesis.
In mitosis, the cell’s nuclear DNA condenses into visible chromosomes and is pulled apart by the mitotic spindle, a specialized structure made of microtubules. Mitosis occurs in four stages: prophase (sometimes divided into early prophase and prometaphase), metaphase, anaphase, and telophase. You can learn more about these stages in the video on mitosis.
In cytokinesis, the cytoplasm of the cell splits in two, forming two new cells. Cytokinesis usually begins just as mitosis ends, with a little overlap. Importantly, cytokinesis is carried out differently in animal and plant cells.
Cytokinesis in animal and plant cells.
- In an animal cell, a contractile ring of cytoskeletal fibres forms in the middle of the cell and contracts inward, producing a cleft called the cleavage furrow. Eventually, the contractile ring pinches the parent cell in two, producing two daughter cells.
- In a plant cell, vesicles derived from the Golgi apparatus move toward the centre of the cell, where they fuse to form a structure called a cell plate. The cell plate expands outward and connects with the cell’s lateral walls, creating a new cell wall that divides the parent cell to form two daughter cells.