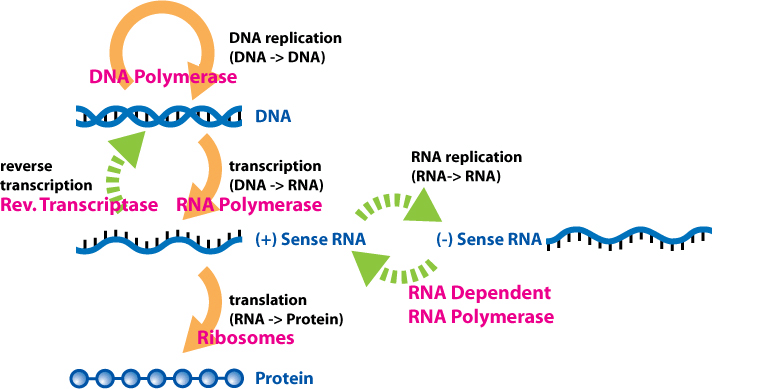
(cDNA) Complementary DNA is a double-stranded DNA version of an mRNA molecule. In higher eukaryotes, an mRNA is a more useful predictor of a polypeptide sequence than a genomic sequence, because the introns have been separated. Researchers prefer to use cDNA over mRNA because RNAs are inherently less stable than DNA and techniques to routinely amplify and purify individual RNA molecules do not exist.
cDNA is made from mRNA with the use of a special enzyme called reverse transcriptase, originally isolated from retroviruses. Using an mRNA molecule as a template, reverse transcriptase synthesizes a single-stranded DNA molecule that can then be used as a template for double-stranded DNA synthesis. It is not necessary to cut the cDNA in order to clone it.
DNA: the building block of life
Deoxyribonucleic acid (DNA) is the molecule that carries the instructions for all aspects of an organism’s functions, from growth to metabolism to reproduction. In living organisms, most of the DNA resides in tightly coiled structures called chromosomes, located within the nucleus of each cell. DNA is made up of four different building blocks, called nucleotides, each of which is made up of one of the four nitrogenous bases. These are the purines: guanine (G) and adenine (A), and the pyrimidines: thymine (T) and cytosine (C).
These nucleotides are attached to a deoxyribose sugar and can join other deoxyribose sugars through phosphate bonds to form long chains, some of which can be more than 100,000,000 molecules long. Since each deoxyribose in a DNA strand is attached to one of four nitrogenous bases (G, A, T, or C), these long chains can carry information.
Groups of three nucleotides form the smallest but most well-defined “words” in the language of DNA. These “words” are called codons. Codons are used to request the joining of specific amino acids to form proteins. For example, the adenosine-adenosine-guanosine (AAG) codon requires the amino acid lysine (Lys) to be incorporated into a protein molecule. The AGG codon calls the amino acid arginine (arg). So AAG-AGG would require a lys to be coupled to an arg in a growing protein chain. There are also codons that, under the right circumstances, require a protein to begin to form (start codons) or a protein chain to end (stop codons). As you can see from this simple example, DNA can carry a huge amount of information.
What is genomic and complementary DNA?
The DNA that resides on the chromosomes inside the nucleus, with all the biological information that will be transferred to the next generation, is called genomic DNA (gDNA). The words “genome” and “genomic” come from the word “gene”. A gene is a set of codons that specify a specific protein chain, along with associated start and stop codons. The word genome is an extension of this concept and means the collection of all the genes and other information contained within the nuclei of the cells of an organism. Often when the word “DNA” is used without further clarification, it refers to gDNA.
In nature, the process for information to be transmitted from DNA can occur through gene replication or gene expression. There are some important factors to keep in mind:
- DNA can copy itself in a process known as replication, using DNA polymerase.
- Information from DNA passes through messenger RNA (mRNA), which contains sets of four nucleotides (uracil, adenine, guanine, and cytosine).
- mRNA is produced when enzymes, such as RNA polymerase, bind to specific genes and copy their information into RNA using ribose sugar (not deoxyribose as in DNA). This process is called transcription.
- Ribosomes assemble around the mRNA, creating a chain of amino acids to create specific proteins. This is called translation.
- Due to the ribose sugar chains, mRNA is short-lived. It is designed to transmit information from the chromosomes in the nucleus to the machinery that makes proteins.
- The mRNA rapidly degrades after it has completed its purpose.
Initially, it was observed that gDNA was always read and transcribed into mRNA, which guided protein formation, and was then removed. The notion that information can always flow from DNA to RNA to protein was jokingly called the central dogma of molecular biology.

The functions of gDNA and cDNA
cDNA can be described as gDNA without all the necessary non-coding regions, thus it gets its name as complementary DNA. The main distinction to be made between cDNA and gDNA is the existence of introns and exons. Introns are nucleotides in genes that do not have coding sequences. Introns are usually cleaved or “removed” from RNA in the transcription process before proteins are created. It should be noted that prokaryotes are not capable of splicing introns. Exons are a necessary part of the coding system and are retained after introns are spliced. Extron’s are non-splicing introns, even though they do not contain coding sequences.
When scientists use viral enzymes to make cDNA from RNA isolated from the cells and tissues they are studying, it does not contain introns because it is spliced into mRNA. The cDNA also does not contain any other gDNA that does not directly code for a protein (referred to as non-coding DNA). Lastly, not all genes in gDNA are transcribed into mRNA at any given time. As a result, the cDNA will only contain genes that are actively being used by a specific cell or tissue at any given time. There is much less total information in cDNA than in gDNA, but the information that remains may be much more relevant to what a researcher is looking for since it does not contain sequences that are unnecessary for DNA function and replication.
Once isolated, gDNA can be used to create genomic libraries for DNA sequencing, fingerprinting, differentiation, and other applications in both the clinical and research fields. cDNA can also be used to make cDNA libraries, permanent collections of cDNA that can be copied and/or stored long-term and is commonly used to clone eukaryotic genes into a prokaryote. In this way, a protein expressed in a eukaryotic organism can be introduced into a prokaryote. For this process, cDNA over gDNA is used, since prokaryotes cannot stimulate introns contained in gDNA.
To isolate cDNA, RNA must first be isolated from an organism. Then, using a reverse transcriptase enzyme, cDNA can be produced. This is the process that retroviruses use to incorporate themselves into the cells of their host. Retroviruses, such as simian immunodeficiency virus (SIV) and avian myeloblastosis virus (AMV), use their cDNA to produce mRNA in the host, leading to the production of viral proteins. This is possible because retroviruses use RNA as genomic material instead of DNA, and it is reverse transcribed into cDNA, which then undergoes normal transcription and leads to viral protein in the host.
Custom and pre-made gDNA and cDNA available on BioChain
BioChain provides access to a comprehensive and well-documented tissue bank containing isolated samples that have been tested for contaminants. As part of rigorous quality control, gDNA samples are analyzed by spectrophotometer and electrophoresis, with concentration determined by UV260 measurement and plant concentration determined by green peak measurement. All gDNA is treated with RNase to remove all RNA.
Genomic DNA comes from unique sources, including hundreds of healthy or diseased organ tissues from humans, animals, and plants. gDNA has applications ranging from SNP analysis, methylation studies, copy number variation (CNV) analysis, comparative genomic hybridization (CGH), Southern blotting, next-generation sequencing, and PCR.