Olfaction is the response to odors and is mediated by a class of membrane-sure proteins known as olfactory receptors (ORs).
An understanding of these receptors serves as a good mannequin for primary sign transduction mechanisms and additionally gives essential clues for the methods adopted by organisms for their final survival utilizing chemosensory notion in search of meals or protection in opposition to predators.
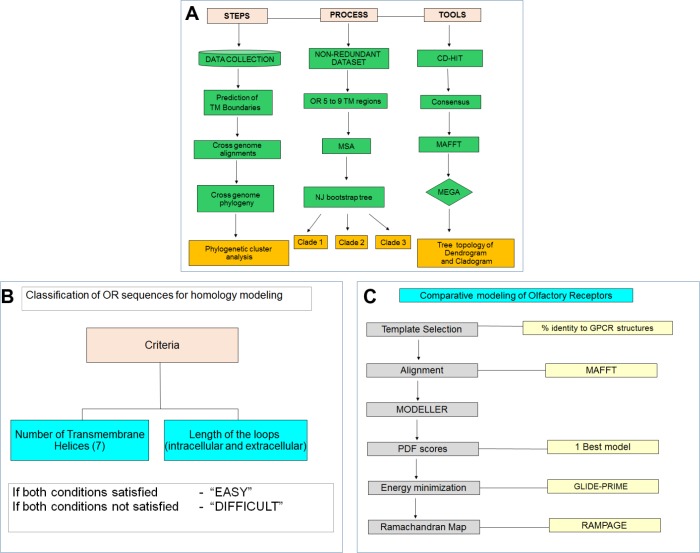
Prior analysis on cross-genome phylogenetic analyses from our group motivated the addressal of conserved evolutionary traits, clustering, and ortholog prediction of ORs.
The database of olfactory receptors (DOR) is a repository that gives sequence and structural info on ORs of chosen organisms (akin to Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, Mus musculus, and Homo sapiens).
Users can obtain OR sequences, examine predicted membrane topology, and receive cross-genome sequence alignments and phylogeny, together with three-dimensional (3D) structural fashions of 100 chosen ORs and their predicted dimer interfaces.
Inbred Strain Variant Database (ISVdb): A Repository for Probabilistically Informed Sequence Differences Among the Collaborative Cross Strains and Their Founders.
The Collaborative Cross (CC) is a panel of just lately established multiparental recombinant inbred mouse strains.
For the CC, as for any multiparental inhabitants (MPP), efficient experimental design and evaluation profit from detailed data of the genetic variations between strains. Such variations might be instantly decided by sequencing, however till now entire-genome sequencing was not publicly obtainable for particular person CC strains. An various and complementary strategy is to deduce genetic variations by combining two items of info: probabilistic estimates of the CC haplotype mosaic from a customized genotyping array, and probabilistic variant calls from sequencing of the CC founders.
The computation for this inference, particularly when carried out genome-huge, might be intricate and time-consuming, requiring the researcher to generate nontrivial and doubtlessly error-susceptible scripts.
To present standardized, simple-to-entry CC sequence info, we now have developed the Inbred Strain Variant Database (ISVdb).
The ISVdb gives, for all of the exonic variants from the Sanger Institute mouse sequencing dataset, direct sequence info for CC founders and, critically, the imputed sequence info for CC strains.
Notably, the ISVdb additionally: (1) gives predicted variant consequence metadata; (2) permits fast simulation of F1 populations; and (3) preserves imputation uncertainty, which can permit imputed knowledge to be refined in the longer term as further sequencing and genotyping knowledge are collected. .

