Simple sequence repeats (SSRs) are areas in DNA sequence that comprise repeating motifs of size 1-6 nucleotides. These repeats are ubiquitously current and are present in each coding and non-coding areas of genome.
A complete of 534 full chloroplast genome sequences (as on 18 September 2014) of Viridiplantae can be found at NCBI organelle genome useful resource.
It gives alternative to mine these genomes for the detection of SSRs and retailer them within the kind of a database. In an try and correctly handle and retrieve chloroplastic SSRs, we designed ChloroSSRdb which is a relational database developed utilizing SQL server 2008 and accessed by ASP.NET.
It gives data of all of the three varieties (perfect, imperfect and compound) of SSRs. At current, ChloroSSRdb comprises 124 430 mined SSRs, with majority mendacity in non-coding area.
Out of these, PCR primers have been designed for 118 249 SSRs. Tetranucleotide repeats (47 079) have been discovered to be probably the most frequent repeat sort, whereas hexanucleotide repeats (6414) being the least ample.
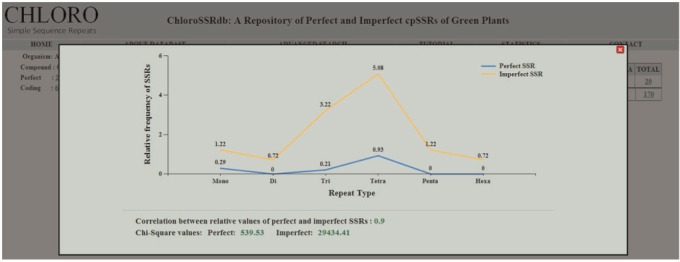
Additionally, in every species statistical analyses have been carried out to calculate relative frequency, correlation coefficient and chi-square statistics of perfect and imperfect SSRs. In accordance with the rising curiosity in SSR research, ChloroSSRdb will show to be a helpful useful resource in growing genetic markers, phylogenetic evaluation, genetic mapping, and many others. Moreover, it would function a prepared reference for mined SSRs in out there chloroplast genomes of green vegetation.
SpliceProt: a protein sequence repository of predicted human splice variants.
The mechanism of various splicing within the transcriptome might improve the proteome range in eukaryotes. In proteomics, a number of research goal to make use of protein sequence repositories to annotate MS experiments or to detect differentially expressed proteins.
However, the out there protein sequence repositories should not designed to totally detect protein isoforms derived from mRNA splice variants. To foster data for the sphere, right here we introduce SpliceProt, a new protein sequence repository of transcriptome experimental knowledge used to research for putative splice variants in human proteomes. Current model of SpliceProt comprises 159 719 non-redundant putative polypeptide sequences.
The evaluation of the potential of SpliceProt in detecting new protein isoforms ensuing from various splicing was carried out through the use of publicly out there proteomics knowledge. We detected 173 peptides hypothetically derived from splice variants, which 54 of them should not current in UniprotKB/TrEMBL sequence repository.
In comparability to different protein sequence repositories, SpliceProt comprises a higher quantity of distinctive peptides and is ready to detect extra splice variants. Therefore, SpliceProt gives a resolution for the annotation of proteomics experiments relating to splice isofoms.

